── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.1 ✔ tibble 3.2.1
✔ lubridate 1.9.4 ✔ tidyr 1.3.1
✔ purrr 1.0.4
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsData transformations
Lecture 9
2025-03-17
Warm up
Setup
Introduction
Prerequisites
- Numeric vectors are the backbone of data science
- But we still need the tidyverse because we’ll use these base R functions inside of tidyverse functions like
mutate()andfilter().
Making numbers
In most cases, you’ll get numbers already recorded in one of R’s numeric types:
integer
double.
In some cases, however, you’ll encounter them as strings, possibly because you’ve created them by pivoting from column headers or because something has gone wrong in your data import process.
Parsing numbers
readr provides two useful functions for parsing strings into numbers: parse_double() and parse_number(). Use parse_double() when you have numbers that have been written as strings:
Use parse_number() when the string contains non-numeric text that you want to ignore. This is particularly useful for currency data and percentages:
Counts
The dplyr::count() is great for quick exploration and checks during analysis:
Counts
If you want to see the most common values, add sort = TRUE:
# A tibble: 105 × 2
dest n
<chr> <int>
1 ORD 17283
2 ATL 17215
3 LAX 16174
4 BOS 15508
5 MCO 14082
6 CLT 14064
7 SFO 13331
8 FLL 12055
9 MIA 11728
10 DCA 9705
# ℹ 95 more rowsIf you want to see all the values:
|> View()|> print(n = Inf).
Counts alternative
Same computation “by hand” with group_by(), summarize() and n().
# A tibble: 105 × 3
dest n delay
<chr> <int> <dbl>
1 ABQ 254 4.38
2 ACK 265 4.85
3 ALB 439 14.4
4 ANC 8 -2.5
5 ATL 17215 11.3
6 AUS 2439 6.02
7 AVL 275 8.00
8 BDL 443 7.05
9 BGR 375 8.03
10 BHM 297 16.9
# ℹ 95 more rowsCount distinct
There are a couple of variants of n() and count() that you might find useful:
n_distinct(x)counts the number of distinct (unique) values of one or more variables. For example, we could figure out which destinations are served by the most carriers:
Weighted count
A weighted count is a sum. For example you could “count” the number of miles each plane flew:
Weighted count
Weighted counts are a common problem so count() has a wt argument that does the same thing:
Counts missing
You can count missing values by combining
sum()andis.na(). In theflightsdataset this represents flights that are cancelled:
Numeric transformations
Transformation functions work well with mutate() because their output is the same length as the input.
Minimum and maximum
The arithmetic functions work with pairs of variables (or columns). Two closely related functions are pmin() and pmax(), which when given two or more variables will return the smallest or largest value in each row:
Minimum and maximum
Note that these are different to the summary functions min() and max() which take multiple observations and return a single value. You can tell that you’ve used the wrong form when all the minimums and all the maximums have the same value:
Modular arithmetic
Modular arithmetic is the technical name for the type of math you did before you learned about decimal places, i.e. division that yields a whole number and a remainder. In R, %/% does integer division and %% computes the remainder:
Modular arithmetic
Modular arithmetic is handy for the flights dataset, because we can use it to unpack the sched_dep_time variable into hour and minute:
Modular arithmetic
We can combine that with the mean(is.na(x)) trick from ?@sec-logical-summaries to see how the proportion of cancelled flights varies over the course of the day. The results are shown in Figure 1.
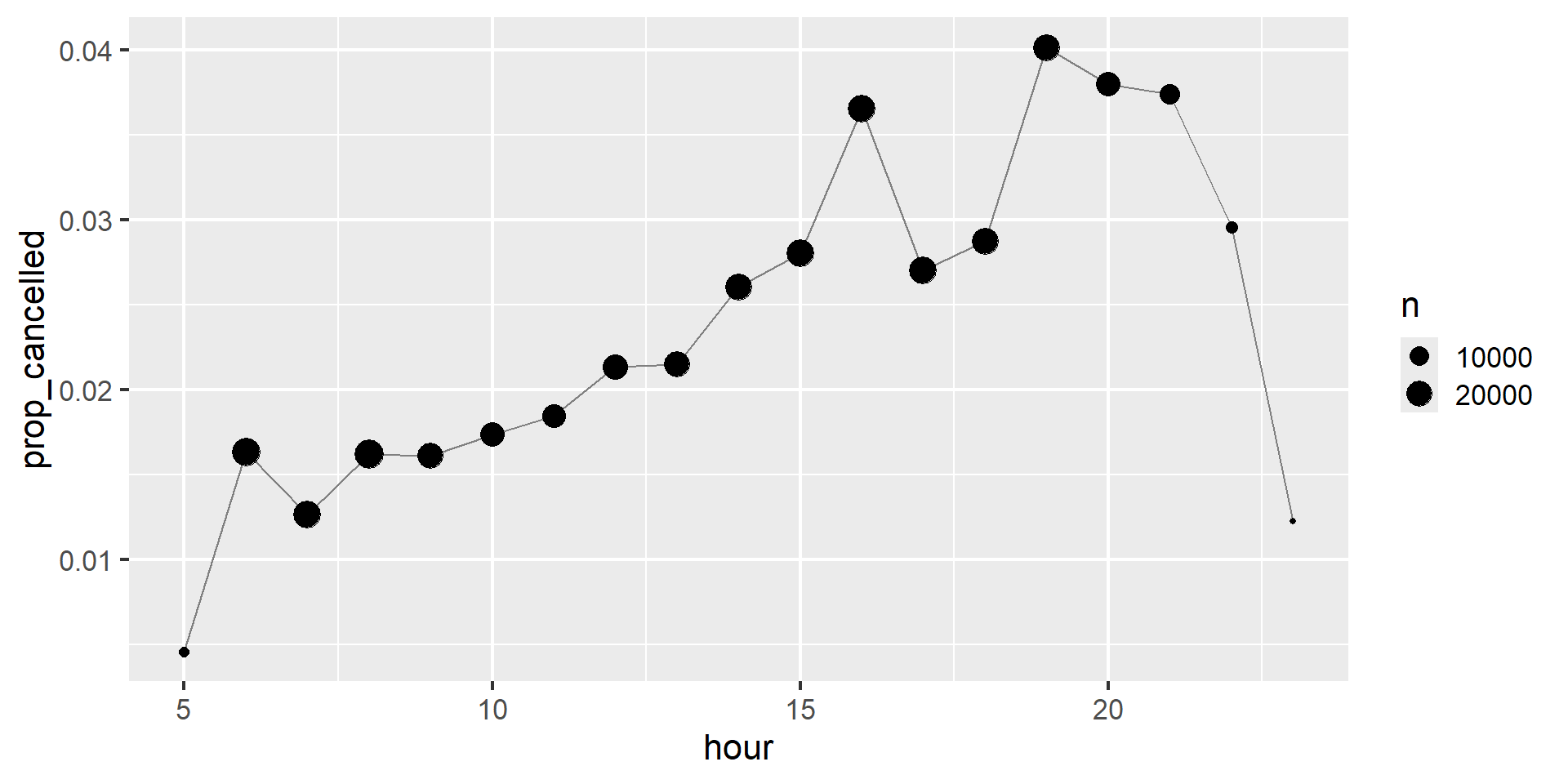
Figure 1: A line plot with scheduled departure hour on the x-axis, and proportion of cancelled flights on the y-axis. Cancellations seem to accumulate over the course of the day until 8pm, very late flights are much less likely to be cancelled.
Logarithms
Three logarithms
In R, you have a choice of three logarithms:
log()(the natural log, base e),log2()(base 2),log10()(base 10).
Logarithm recommendation
We recommend using log2() or log10():
log2()is easy to interpret because a difference of 1 on the log scale corresponds to doubling on the original scale and a difference of -1 corresponds to halving;log10()is easy to back-transform because (e.g.) 3 is 10^3 = 1000.
The inverse of log() is exp(); to compute the inverse of log2() or log10() you’ll need to use 2^ or 10^.
Rounding
Use round(x) to round a number to the nearest integer:
Rounding digits
You can control the precision of the rounding with the second argument, digits. round(x, digits) rounds to the nearest 10^-n so digits = 2 will round to the nearest 0.01. This definition is useful because it implies round(x, -3) will round to the nearest thousand, which indeed it does:
Rounding digits - weirdness
There’s one weirdness with round() that seems surprising at first glance:
round() uses what’s known as “round half to even” or Banker’s rounding: if a number is half way between two integers, it will be rounded to the even integer. This is a good strategy because it keeps the rounding unbiased: half of all 0.5s are rounded up, and half are rounded down.
Rounding digits - floor/ceiling
round() is paired with floor() which always rounds down and ceiling() which always rounds up:
Cutting numbers into ranges
Use cut()1 to break up (aka bin) a numeric vector into discrete buckets:
Cutting numbers into ranges
The breaks don’t need to be evenly spaced:
Cutting numbers into ranges
You can optionally supply your own labels. Note that there should be one less labels than breaks.
Cutting numbers into ranges
Any values outside of the range of the breaks will become NA:
[1] <NA> <NA> (0,5] (5,10] <NA>
Levels: (0,5] (5,10] (10,15] (15,20]See the documentation for other useful arguments like right and include.lowest, which control if the intervals are [a, b) or (a, b] and if the lowest interval should be [a, b].
Cumulative and rolling aggregates
Base R provides cumsum(), cumprod(), cummin(), cummax() for running, or cumulative, sums, products, mins and maxes. dplyr provides cummean() for cumulative means. Cumulative sums tend to come up the most in practice:
If you need more complex rolling or sliding aggregates, try the slider package.
General transformations
The following sections describe some general transformations which are often used with numeric vectors, but can be applied to all other column types.
Ranks
dplyr provides a number of ranking functions inspired by SQL, but you should always start with dplyr::min_rank(). It uses the typical method for dealing with ties, e.g., 1st, 2nd, 2nd, 4th.
Ranks
Note that the smallest values get the lowest ranks; use desc(x) to give the largest values the smallest ranks:
Ranks alternatives
If min_rank() doesn’t do what you need, look at the variants dplyr::row_number(), dplyr::dense_rank(), dplyr::percent_rank(), and dplyr::cume_dist(). See the documentation for details.
df <- tibble(x = x)
df |>
mutate(
row_number = row_number(x),
dense_rank = dense_rank(x),
percent_rank = percent_rank(x),
cume_dist = cume_dist(x)
)# A tibble: 8 × 5
x row_number dense_rank percent_rank cume_dist
<dbl> <int> <int> <dbl> <dbl>
1 1 1 1 0 0.143
2 3 4 3 0.5 0.571
3 2 2 2 0.167 0.429
4 2 3 2 0.167 0.429
5 4 5 4 0.667 0.714
6 20 7 6 1 1
7 15 6 5 0.833 0.857
8 NA NA NA NA NA Offsets
dplyr::lead() and dplyr::lag() allow you to refer to the values just before or just after the “current” value. They return a vector of the same length as the input, padded with NAs at the start or end:
Offsets - lag
x - lag(x)gives you the difference between the current and previous value.x == lag(x)tells you when the current value changes.
You can lead or lag by more than one position by using the second argument, n.
Consecutive identifiers
Sometimes you want to start a new group every time some event occurs. For example, when you’re looking at website data, it’s common to want to break up events into sessions, where you begin a new session after a gap of more than x minutes since the last activity. For example, imagine you have the times when someone visited a website:
Consecutive identifiers
And you’ve computed the time between each event, and figured out if there’s a gap that’s big enough to qualify:
events <- events |>
mutate(
diff = time - lag(time, default = first(time)),
has_gap = diff >= 5
)
events# A tibble: 14 × 3
time diff has_gap
<dbl> <dbl> <lgl>
1 0 0 FALSE
2 1 1 FALSE
3 2 1 FALSE
4 3 1 FALSE
5 5 2 FALSE
6 10 5 TRUE
7 12 2 FALSE
8 15 3 FALSE
9 17 2 FALSE
10 19 2 FALSE
11 20 1 FALSE
12 27 7 TRUE
13 28 1 FALSE
14 30 2 FALSE Consecutive identifiers
But how do we go from that logical vector to something that we can group_by()? cumsum(), from Section 5.11, comes to the rescue as gap, i.e. has_gap is TRUE, will increment group by one (?@sec-numeric-summaries-of-logicals):
# A tibble: 14 × 4
time diff has_gap group
<dbl> <dbl> <lgl> <int>
1 0 0 FALSE 0
2 1 1 FALSE 0
3 2 1 FALSE 0
4 3 1 FALSE 0
5 5 2 FALSE 0
6 10 5 TRUE 1
7 12 2 FALSE 1
8 15 3 FALSE 1
9 17 2 FALSE 1
10 19 2 FALSE 1
11 20 1 FALSE 1
12 27 7 TRUE 2
13 28 1 FALSE 2
14 30 2 FALSE 2Consecutive identifiers
Another approach for creating grouping variables is consecutive_id(), which starts a new group every time one of its arguments changes. For example, inspired by this stackoverflow question, imagine you have a data frame with a bunch of repeated values:
Consecutive identifiers
If you want to keep the first row from each repeated x, you could use group_by(), consecutive_id(), and slice_head():
Numeric summaries
Just using the counts, means, and sums that we’ve introduced already can get you a long way, but R provides many other useful summary functions. Here is a selection that you might find useful.
Center
- Function
mean() - Function
median()
Minimum, maximum, and quantiles
min()andmax()will give you the largest and smallest values.- Another powerful tool is
quantile()which is a generalization of the median:quantile(x, 0.25)will find the value ofxthat is greater than 25% of the values,quantile(x, 0.5)is equivalent to the median,quantile(x, 0.95)will find the value that’s greater than 95% of the values.
Minimum, maximum, and quantiles
For the flights data, you might want to look at the 95% quantile of delays rather than the maximum, because it will ignore the 5% of most delayed flights which can be quite extreme.
flights |>
group_by(year, month, day) |>
summarize(
max = max(dep_delay, na.rm = TRUE),
q95 = quantile(dep_delay, 0.95, na.rm = TRUE),
.groups = "drop"
)# A tibble: 365 × 5
year month day max q95
<int> <int> <int> <dbl> <dbl>
1 2013 1 1 853 70.1
2 2013 1 2 379 85
3 2013 1 3 291 68
4 2013 1 4 288 60
5 2013 1 5 327 41
6 2013 1 6 202 51
7 2013 1 7 366 51.6
8 2013 1 8 188 35.3
9 2013 1 9 1301 27.2
10 2013 1 10 1126 31
# ℹ 355 more rowsVariation
Two commonly used summaries are
the standard deviation
sd(x),the inter-quartile range,
IQR().
Positions
There’s one final type of summary that’s useful for numeric vectors, but also works with every other type of value: extracting a value at a specific position:
first(x),last(x), andnth(x, n).
Positions
For example, we can find the first, fifth and last departure for each day:
flights |>
group_by(year, month, day) |>
summarize(
first_dep = first(dep_time, na_rm = TRUE),
fifth_dep = nth(dep_time, 5, na_rm = TRUE),
last_dep = last(dep_time, na_rm = TRUE)
)`summarise()` has grouped output by 'year', 'month'. You can override using the
`.groups` argument.# A tibble: 365 × 6
# Groups: year, month [12]
year month day first_dep fifth_dep last_dep
<int> <int> <int> <int> <int> <int>
1 2013 1 1 517 554 2356
2 2013 1 2 42 535 2354
3 2013 1 3 32 520 2349
4 2013 1 4 25 531 2358
5 2013 1 5 14 534 2357
6 2013 1 6 16 555 2355
7 2013 1 7 49 536 2359
8 2013 1 8 454 544 2351
9 2013 1 9 2 524 2252
10 2013 1 10 3 530 2320
# ℹ 355 more rowsNB: Because dplyr functions use _ to separate components of function and arguments names, these functions use na_rm instead of na.rm.
With mutate()
As the names suggest, the summary functions are typically paired with summarize(). However, because of the recycling rules we discussed in ?@sec-recycling they can also be usefully paired with mutate(), particularly when you want do some sort of group standardization. For example:
x / sum(x)calculates the proportion of a total.(x - mean(x)) / sd(x)computes a Z-score (standardized to mean 0 and sd 1).(x - min(x)) / (max(x) - min(x))standardizes to range [0, 1].x / first(x)computes an index based on the first observation.